One of the highest-priority use cases within PerMedCoE involves the development of high performance computing (HPC)-compatible workflows for patient-specific COVID-19 modeling using single-cell data. To enable such modeling to be executed at multiple temporal and spatial scales, the analytical pipelines involved must be highly scalable and efficient.
Workflow design tasks within PerMedCoE include software development (e.g. enabling support for distributed computing) and packaging analytical tools into containers. The containers are assembled into building blocks aimed at addressing specific analytical functionalities as required by different PerMedCoE use cases, and which can be used by workflow managers including PyCompSs.
To connect these tasks with the COVID-19 use case, work is underway to develop a pilot workflow for single-cell COVID-19 modeling using MaBoSS and PhysiBoSS. Further to building blocks for identifying gene candidates and carrying out single-cell RNA-Seq data processing, functionalities included in the workflow involve parallelised modeling (including within individual analyses) followed by meta-analysis of the resulting data. The entire workflow is designed around a unified command line interface that is being developed to support all PerMedCoE use cases.
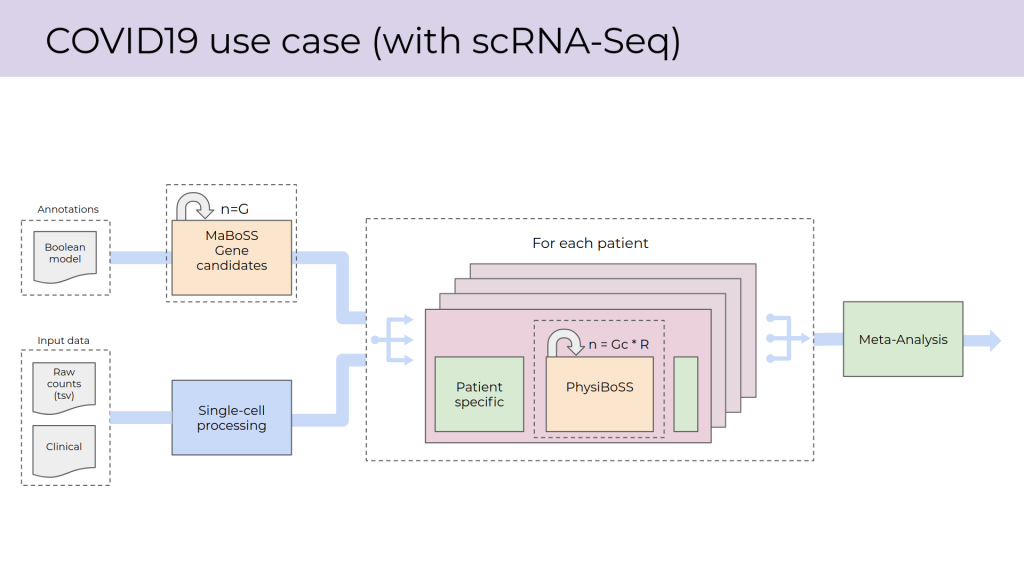
Data types and different analytical steps included in the COVID-19 pilot workflow. Image by José Carbonell (BSC).
The main goals of the COVID-19 pilot workflow are to provide an opportunity for workflow testing on different HPC platforms and to investigate optimal ways to group the functionalities involved into building blocks (e.g. in terms of granularity). The pilot workflow will also serve as the basis for future development activities in relation to the COVID-19 use case. Current development goals include further PhysiBoSS development work and including tools for disease map processing, model exploration (including EMEWS) and trajectory analysis.
Author: Jesse Harrison (CSC – IT Center for Science)





