The emergence of cell resistance in cancer treatment is a complex phenomenon. Studying the interplay between different molecular mechanisms and population-level dynamics is a critical step to understand the emergence and evolution of cell resistances. In this context, multi-scale models are a useful tool to study biology at a very different time and spatial scales, as they can integrate different processes that take place at the molecular, cellular and intercellular levels.
PerMedCoE experts have developed a multi scale model of tumour spheroid to study drug supply strategies in different conditions with the aim to provide novel tools to help to improve cancer treatments. Because of their complexity, multi-scale models require challenging computational simulations to address the result of an in-silico experiments.
With the help of High Performance Computing (HPC), BSC researchers have developed a HPC-based model exploration workflow (see figure below) to efficiently explore the parameter space of effective treatments in a variety of conditions. In collaboration with the Infore project, the implemented workflow was deployed in the MareNostrum 4 supercomputer to run the distributed model exploration allowing in-silico optimized treatment strategies.
Workflow overview
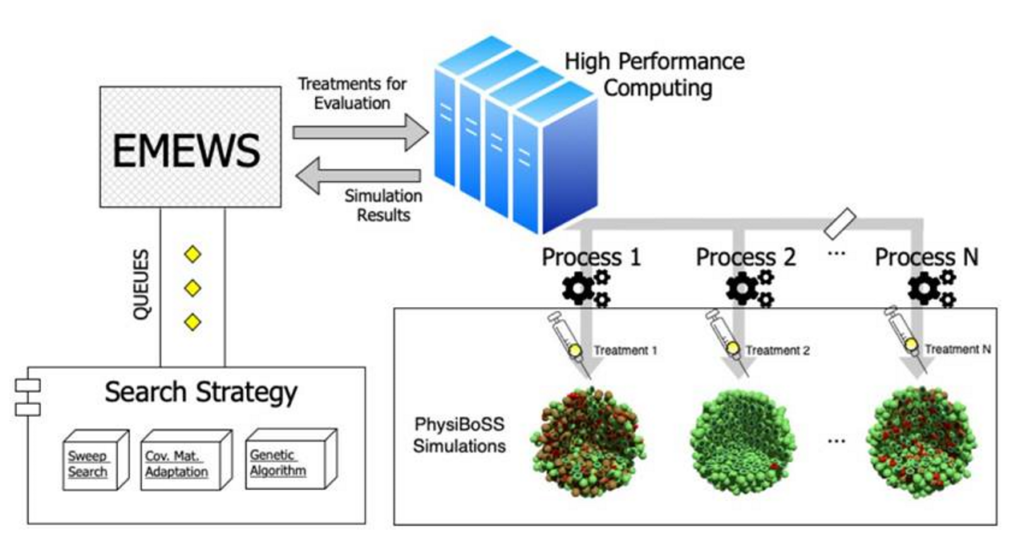
The study is a proof-of-concept that highlights why “multi-scale models are a useful tool to study biology at a very different time and spatial scales, as they can integrate different processes that take place at the molecular, cellular and intercellular levels.”, says Miguel Ponce de León, Senior BSC researcher and first author of the publication. The results of this study have been recently published in the article titled “Optimizing dosage-specific treatments in a multi-scale model of a tumor growth” in the journal Frontiers in Molecular Biosciences. The study shows that cells’ spatial geometry, as well as, population variability should be considered when optimizing treatment strategies in order to find robust parameter sets.
“Bringing together the systems biology and high-performance computing communities is key for the progress of cancer treatments”
Miguel Ponce de León
Thanks to PerMedCoE, significant efforts are being done in this direction bringing closer the systems biology and HPC communities taking take full advantage of HPC clusters. BSC scientists plan to apply the implemented workflow and developed HPC tools to other PerMEdCoE use-cases in the near future.





