Omics data is increasingly available in the fields of healthcare and medicine, and mathematical models that rely on this data constitute a promising approach for exploring and predicting patient drug responses. New modelling methods for predicting drug responses could help clinicians provide efficient treatment for diseases including cancer, diabetes or immune diseases.
Of particular interest from a clinical viewpoint is the development of modelling methods that account for patient heterogeneity in response to treatments, that is, the variation in how patients with the same disease respond to a specific treatment. Capturing this heterogeneity would enable the design of next-generation modelling methods that could be used to recommend personalised drug treatments for individual patients (or groups of patients).
To contribute to this goal and to the PerMedCoE Drug Synergies use case, PerMedCoE partners at Institut Curie and the University of Heidelberg developed a workflow that facilitates the prediction of personalised targeted drug combinations for cancer treatment, using CARNIVAL (Trairathphisan et al. 2021), PROFILE (Béal et al. 2019) and MaBoSS (Stoll et al. 2012, 2017) software. As a proof of concept, the workflow has successfully been run on the BSC MareNostrum 4 supercomputer.
The workflow
The workflow requires two inputs: omics (e.g., transcriptomics or proteomics) data and a Boolean model. The Boolean model can be an existing model that is available in public databases, such as BioModels or Cell Collective, or can be directly inferred from the data. A personalised data integration methodology (PROFILE) is used to generate a mathematical model for each patient or cell line. Single and combined drug treatment simulations are then performed to select the best output. The final result consists of a list of single and combined potential targets for each patient or cell line.
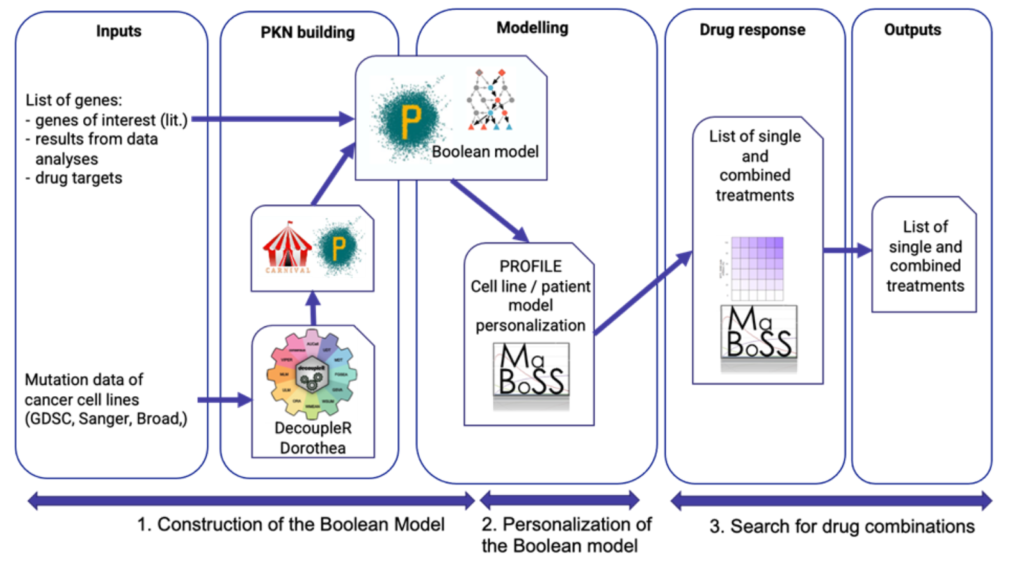
Image by: Laurence Calzone (Institut Curie) and Pablo Rodriguez-Mier (University of Heidelberg)
A Boolean model focused on the early steps of cancer metastasis (published in 2015 by PerMedCoE members at Institut Curie) and the mutation and transcriptomics data of 72 cell lines of colon cancer was the input used to run the workflow on MareNostrum 4. By analysing the data in parallel, it was possible to simulate 72 cell lines on 72 CPU cores in less than 20 minutes. The results included some highly relevant drug combinations that concur with previously published results, thus demonstrating the promise of the pipeline with a concrete biological problem and patient data.
In addition to upscaling the workflow to analyse a greater number of cell lines, researchers are currently involved in an ongoing development task to augment the Drug Synergies workflow with functionalities for drug response (IC50) prediction using machine learning tools, including JAX. The results of this task will be used to further extend the list of potential drug targets identified by the workflow.





