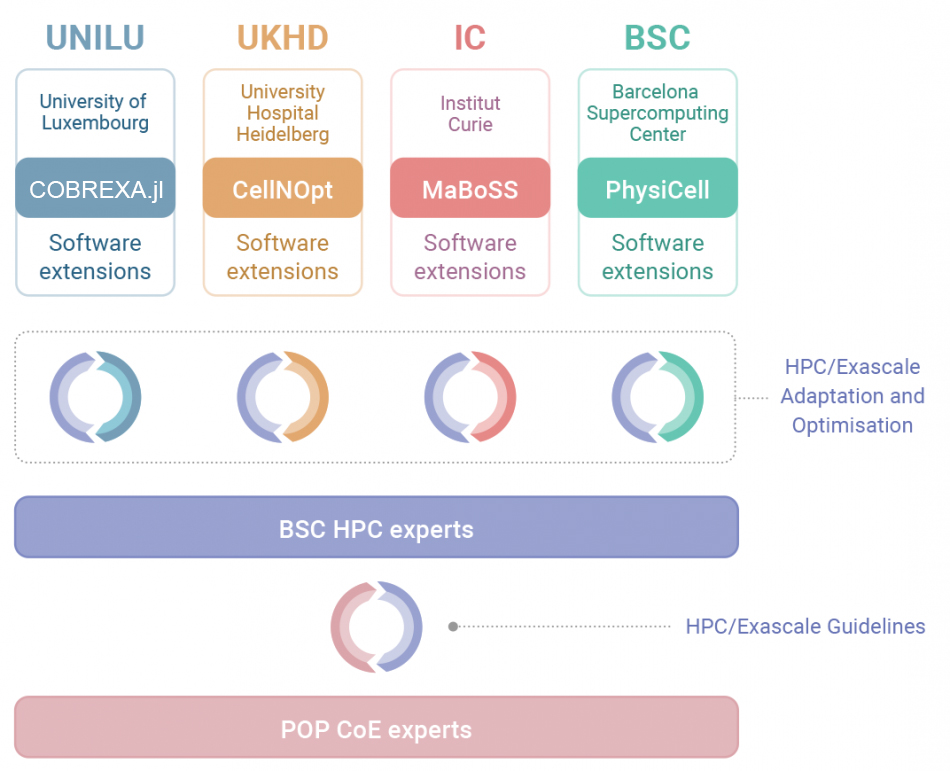
Core Applications
- COBREXA.jl for the simulation of cellular metabolism at genome-scale
- CellNOpt for modelling signal transduction networks
- MaBoSS for stochastic simulations of Boolean models
- PhysiCell an agent-based modelling framework for simulating cell-cell interactions

Constraint-Based Reconstruction and Analysis (COBREXA) is an ever-evolving computational framework used to reconstruct and analyse models of cell metabolism at genome-scale. At the technical level, the framework relies on the introduction of the linear constraints such as mass conservation and thermodynamics to define the so-called flux space (set of reaction fluxes that are biologically feasible). The flux space associated to a metabolic model can be explored using different approaches, such as optimisation methods (e.g. linear programming) or sampling techniques (Monte Carlo methods), to identify biologically relevant solutions (i.e. phenotypes).
COBREXA methods, such as the widely used Flux Balance Analysis (FBA), relies on linear constraints and objective functions and do not require kinetic parameters to identify meaningful metabolic states, which make them highly scalable and thus, the only approach applicable to genome-scale models. As a consequence, this framework has enabled the reconstruction and analysis of many models organisms of scientific relevance, such as E. coli and S. cerevisiae, and the human cell metabolism (starting with RECON1 and currently aiming for a Virtual Metabolic Human) and also whole-reconstructed microbiomes. COBREXA methods have a broad spectrum of biotechnological and biomedical applications including rational design strategies for engineering microorganisms of industrial interest and model guided prediction of biomarkers and gene targets for complex diseases (e.g. cancer, Alzheimer) among many others. For all these reasons, COBRA has become the most-widely adopted computational framework for network reconstruction, integrative analysis of omics data and mechanistic modelling of cell metabolism.
COBREXA implementations are currently highly popular for constraint-based modelling. COBRA methods have been written and distributed in different programming languages such as MatLab, Python and Julia. COBREXA methods are mostly run in desktop environments by scientists, and they hit their applicability limits when running on pre-exascale architectures. To overcome this computational bottleneck, COBREXA.jl exploits the parallel-ready syntax and the availability of powerful libraries for numerical methods of the Julia programming language. Analysis methods implemented in COBREXA.jl transparently and efficiently scale to the pre-exascale infrastructure, enabling the users to work with larger models and much greater amounts of model perturbations than possible before.
Signal transduction networks are a very common way to depict the underlying mechanisms of the biological response to a given stimulus from the environment or a genetic perturbation. These networks usually involve genes, proteins, chemical components and/or phenotypes. These networks can be transformed into models that use different mathematical methodologies to be analysed, such as ordinary differential equations, partial differential equations, Boolean algebra, fuzzy dynamics, discrete event dynamics, etc. Signal transduction models have been widely used in Biology since Jacob and Monod published their seminal paper in 1961 and are a central part of current Systems Biology research and many relevant projects such as the WholeCell Modeling Initiative (e.g., WholeCellKB, from the Karr group at Icahn School of Medicine at Mount Sinai, or E-Cell, project from the Takahashi group at RIKEN).
NEW RELEASES
Official repository
The official repository of COBREXA tool is now available.
How to use the tool documentation
CellNOpt is a software package to create and simulate logic-based models of signal transduction networks using different logical formalisms, including Boolean, Fuzzy, or differential equations. All these capabilities make CellNOpt a powerful and self-contained toolbox for modelling cell signalling networks. While there are several available tools for simulating signalling networks, CellNOpt is particularly focused on simulating large-scale models as well as their training or fitting to experimental data. Training a model is a critical step which implies learning parameters from experimental data by minimising a bipartite optimisation function. The optimisation is performed using a built-in genetic algorithm or a scatter search, approaches that are very suitable to run in HPC environments and that will benefit from exascale capabilities. CellNOpt code is mostly written in R language with a critical piece of the code implemented in C, uses an open license (GNU General Public License, version 2) and was partially developed in the context of the European project BioPreDyn.
Signal transduction networks can also be studied using Boolean algebra, where the sets of discrete variables are represented as abstractions of activity level linked by logical rules as signed interactions. These logic models represent a convenient way to model huge regulatory and signalling networks as they allow integration of literature and heterogeneous data and provide qualitative analysis, which is a good compromise as most of the data available in Biology is semi-quantitative. Logic modelling has been widely used in different projects, such as drug synergies and cancer studies.
NEW RELEASES
How to use the tool documentation
Illustrative use case
MaBoSS is open-source software for simulating the dynamics of logic models representing gene regulatory networks or signalling pathways. MaBoSS approach to simulate the state of a system relies on a continuous-time Markov process (also termed Gillespie algorithm) on the state transition graph induced by a given Boolean model and allows estimating the probability distribution of stationary states (attractors) in Boolean models. MaBoSS code is coded in C++ and C language and allows parallel computing using a multi-thread implementation based on OpenMP. For this reason, MaBoSS exhibits a high performance when compared to other available options such as GinSIM which was designed for running in desktop computers through a graphical user interface. However, constructing the transition graph and estimating the distribution of stationary states for large model is still challenging due the high-memory demands of simulations. Thus, to improve the runtimes of these high-memory-demanding simulations a redesign to exploit the advantage of heterogeneous platforms’ (i.e. combining CPUs with GPUs) and heterogeneous memory architectures will be needed. The research leading to the development of MaBoSS has received funding from the European projects APO-SYS and ASSET.
CellNOpt and MaBoSS have been selected to address the logical modelling approach for two reasons:
- they address quantitative issues of logical modelling;
- they are part of the well-established CoLoMoTo consortium, which facilitates the use and communication between tools.
This provides flexibility with regards to the intracellular models that can be used and the comparison of their outputs.
Agent-based (or cell-based) models have been used to simulate individual cells as well as collective behaviours within tissue environments. These models, especially lattice-free centre-based models, have been increasingly used in Systems Biology to link cancer development to microenvironment conditions. Centre-based models track each cell’s centre of mass or volume, typically by using a single agent per cell that represents cells as points or cell volumes. Agent-based modelling has been used to tackle biological problems such as the role of hypoxia in cancer or drug regimes’ studies and also in large consortia such as DARPA’s XAI and NIH’s HubMAP.
NEW RELEASES
Official Repository
The new version of MaBoSS is already available at the official repository. The repository includes:
- Latest MaBoSS version
- Documentation to learn how to use the tool
Additional resources published
- Phyton bindings of MaBoSS
- New web interface (to be deployed locally using docker or freely accesible)
PhysiCell is an open-source and powerful lattice-free agent-based framework for multi-scale simulation of multicellular systems. It was designed to simulate the evolution and the dynamics of a population of cells such as tumours. PhysiCell is a very flexible framework that is fully written in C++, and relies on a very efficient solver of partial differential equation (BioFVM) to simulate the microenvironment, and a built-in mechanics solver to simulate cell-cell and cell-environment mechanical interactions. An extension of PhysiCell (PhysiBoSS25) allows the integration of an individual Boolean model for the signalling networks embedded into each cell-agent. The main advantages that PhysiCell exhibits over other mature frameworks, such as Chaste, are that it is a light-weight, very efficient and self-contained framework, that is very extensible, and it supports standard formats including MutiCellDS. While the development and maintenance of PhysiCell is led by Indiana University, the PhysiBoSS extension was developed at IC and received funding from the PrECISE project.
NEW RELEASES
Official Repository
The official repository of the new PhyCell-X release is now available. The repository includes:
- Source code
- Getting started tutorial: for execution on a single desktop machine & for execution on a professional HPC environment with Slurm job management system
Illustrative examples
Learn how to design simulations in a wide variety of biological scenarios with our illustrative examples.