
PerMedCoE webinars are open to everyone interested in PerMedCoE tools and activities. The webinars will include a 30-40 minutes presentation and a Q&A section of around 15 minutes. The recording of this webinar will be publicly available on this web page and the PerMedCoE YouTube channel.
Webinar:
| Speakers: | Viviam Bermúdez (Norwegian University of Science and Technology), Marco Fariñas (Norwegian University of Science and Technology), and Malvina Marku (INSERM Toulouse Cancer Research Center) |
| Date and time: | Thursday 05 October 2023, 15-16 CEST |
| Target groups: | Bioinformaticians Researchers in the life sciences Biomedical scientists Anyone interested in simulation of intercellular interactions |
| Learning outcomes: | Describe how Boolean models can be customised into patient-specific models Describe data-driven methodology to investigate cancer cell – host cell interactions |
| Watch the recording | |
| Access the slides |
Viviam Bermúdez
Towards Tailored Cancer Therapies: Robust platform for drug testing in patient-specific Boolean models
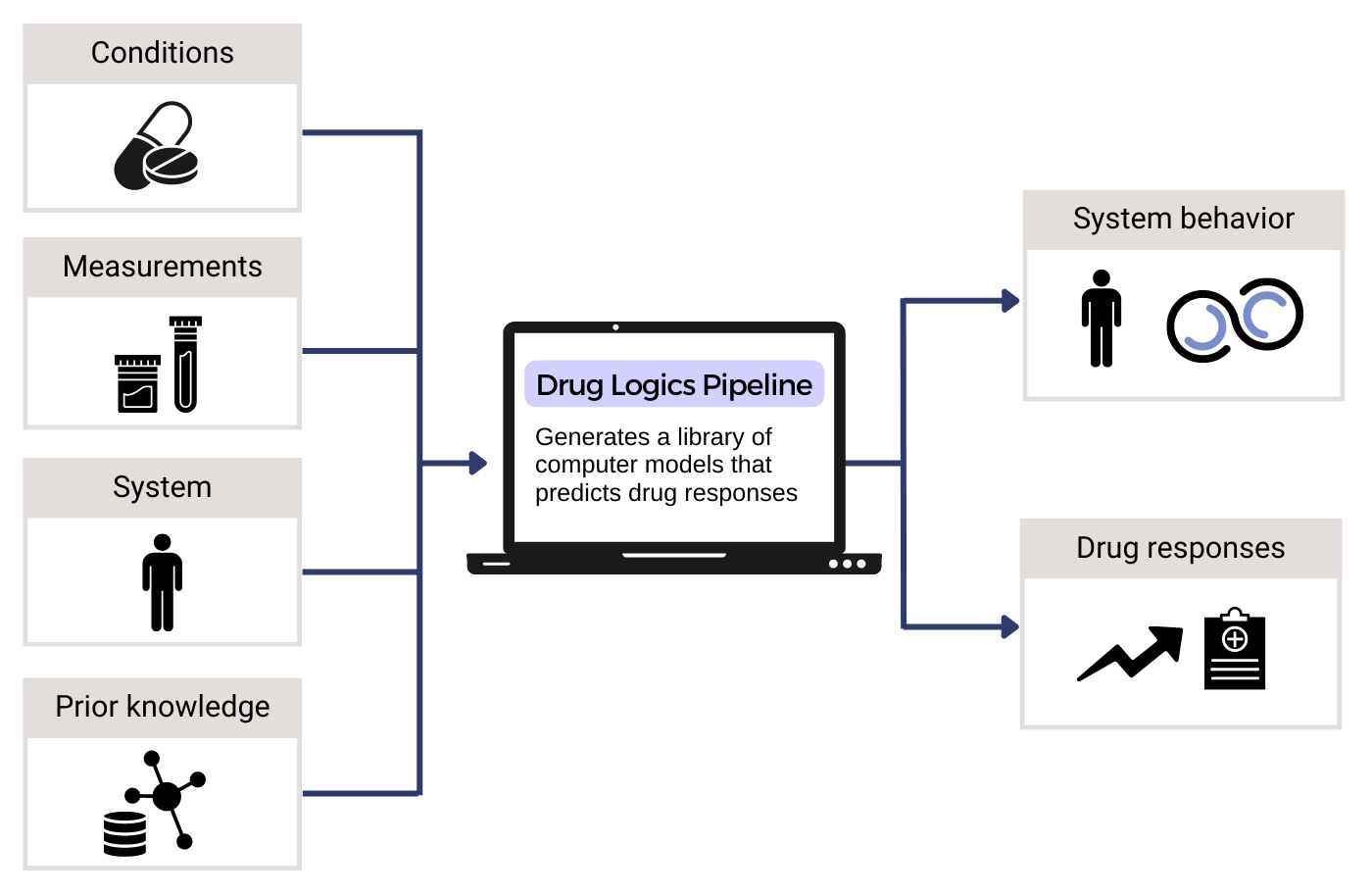
Functional precision medicine offers a crucial opportunity in oncology, allowing treatment design and testing in patient companion models. However, the key challenge lies in developing efficient computer models to select optimal candidate therapies that can be tested in limited patient-derived material. Our project aims to develop a platform that enables the customization of Boolean models into patient-specific models to simulate the action of candidate drugs. We employ bioinformatics, artificial intelligence, and machine learning approaches to integrate and harmonise datasets on available drugs, drug targets, existing drug synergy data, and precise activity states of key regulatory network proteins derived from patient omics data. The model’s predictive accuracy for drug responses enables in vitro testing and data-driven decisions for tailored treatment plans, considering a wide range of treatment designs and approved drugs, including repurposed and novel combinations. Through the fusion of diverse biological data, model simulations, and prediction analysis, the platform serves as a tool within the NTNU DrugLogics pipeline, paving the way in the field of precision oncology.

Viviam Bermúdez is a PhD candidate at the Norwegian University of Science and Technology, specialising in Boolean models. Her academic journey began in general biology back in her home country, Venezuela, at Simón Bolívar University. Then, she hopped over to Norway for a one-year student exchange, where she unexpectedly found herself staying longer to complete a master’s degree in molecular cell biology – diving deep into systems biology and cell modelling. Now, in her Computational Cancer Biology PhD, she combines machine learning and modelling to address complex challenges of precision medicine. Beyond her coding skills, she also has green fingers, so she finds peace caring for her plants during her free time. Looking ahead, she aspires to contribute to the field of precision medicine through her PhD project and continue exploring the fascinating world of “digital-twins” – without forgetting the ethical side of things.
Marco Fariñas
Computational modelling of High Grade Serous Ovarian Cancer (HGSOC)
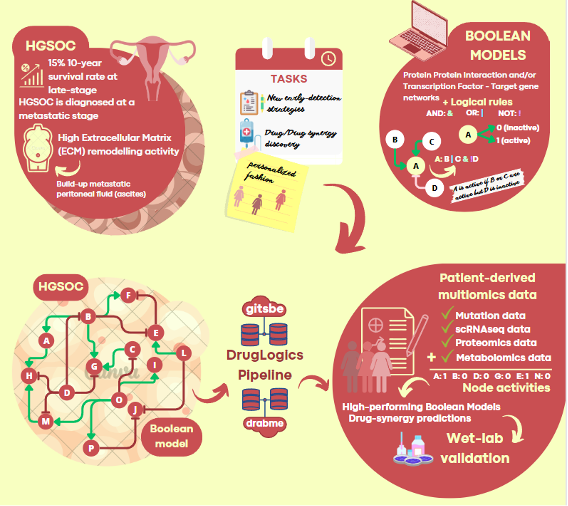
High Grade Serous Ovarian Cancer is a lethal gynaecological malignancy with progressively resistant metastases. To date, our results have revealed mechanisms by which the tumour micro-environment (TME) modulates the dynamical tumour behaviour. In this context, we have developed a HGSOC Boolean network that includes most relevant pathways to disease progression. By usage of multiomics data (focusing on mutation and transcriptomics-derived transcription-factor-activity data) generated upon different experimental designs, the model will be trained to capture the influence of crosstalk that takes place between tumour cells and stromal cells and the extracellular matrix. This will ultimately provide a powerful resource to uncover the molecular mechanisms behind HGSOC aggressiveness. The subsequent ability to use cohort- and patient-specific molecular profiles for treatment predictions will be of immediate clinical relevance. Methodologically, the project design will provide a user-friendly reproducible pipeline that can be easily adapted and replicated for similar and different studies.

Originally from Ponferrada, in north-western Spain, Marco Fariñas is an enthusiastic nature lover who enjoys spending free time outdoors. His academic journey began with a Bachelor’s degree in Biology at the Complutense University of Madrid. Following his interest for cellular and molecular biology, he moved to Trondheim, Norway, where he pursued a Master’s degree at the Norwegian University of Science and Technology (NTNU). During this period, Marco did his thesis focusing on Boolean modelling of macrophage polarisation and its relevance to acute COVID-19. This research was conducted under the guidance of Martin Kuiper within the DrugLogics group. Following the completion of the Master’s program, Marco moved to Barcelona and joined Marta Cascante’s research group, at Barcelona University. There, he delved into the intricate metabolic alterations triggered by SARS-CoV-2 infection. In his latest pursuit, Marco returned to Trondheim to start his PhD journey at NTNU, under the supervision of Kaisa Lehti. His research leverages Boolean modelling to explore the dynamics of the tumour microenvironment and its influence on the aggressiveness of high-grade serous ovarian cancer.
Malvina Marku
Dissecting cellular communication in the tumour microenvironment through multiscale dynamical modelling
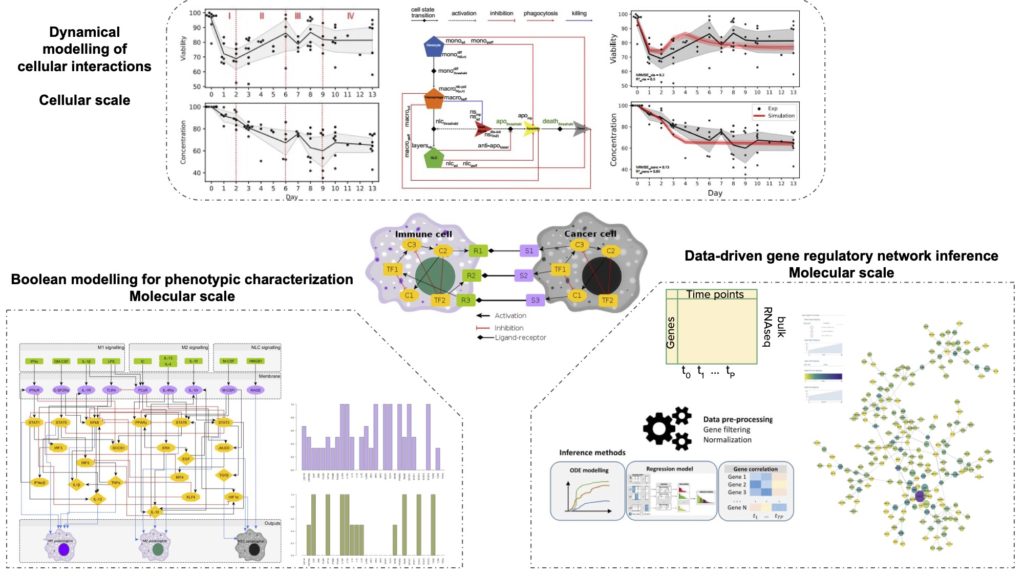
The tumour microenvironment (TME) can be seen as a complex system containing multiple cell types interacting through contact and cytokine exchanges. In particular, immune cells play a major role in cancer development and their characterization allows a better understanding of the TME. In this context, network approaches and mathematical modelling allow studying interactions between immune and cancer cells to obtain relevant information about cellular reprogramming and state transitions, and identify novel molecular interactions and potential drug targets. This project aims to investigate how regulatory interactions between genes characterise the cellular behaviour of cancer cells in the presence of immune cells at the molecular and cellular level. Prior work in our lab has characterised the functional processes determining cancer cell – immune cell interactions at the cellular level, revealing important biological processes determining cancer cell survival. Then, following a data-driven gene regulatory network inference on cancer cells, we identified novel transcription factors involved in cancer cell – immune cell crosstalk, thus better understanding their mechanisms of survival. Integrating these two scales, we aim to recapitulate the processes that lead to the formation of pro-tumor phenotypes of immune cells, specifically highlighting their effect on cancer cell survival.

Malvina Marku studied her undergraduate degree in Physics at the University of Tirana, Albania, where she pursued her MSci project in the field of Nonlinear Dynamics, studying the qualitatively universal behaviour of recursive systems. As a PhD student on Computational Physics at the University of Tirana, her PhD thesis focused on mathematical modelling of regulatory networks and inference methods of network reaction rates. She joined Pancaldi’s lab at the Cancer Research Center of Toulouse as a postdoc where she is currently working on multi-scale dynamical modelling of the tumour microenvironment. The research is focused on Chronic Lymphocytic Leukaemia (CLL) microenvironment, and the crucial role of the cancer cell-immune cell interaction in defining the cancer cell survival. Her main interest is in applying dynamical modelling and networks theory to study the dynamics and emerging behaviour of complex biological systems, and to gain a deeper understanding of systems’ evolution from the molecular network perspective. She is interested in using computational models that are able to capture emerging biological behaviour and evolving patterns in spatially structured tumours for enabling more accurate and clinically relevant predictions regarding disease course and treatment outcome.





