PerMedCoE, is working to scale up various computational biology tools to leverage the next generation of high-performance computing exascale platforms. One promising framework for this is agent-based modelling, which can be used to create digital twins that can act as clinical decision tools. Given the diversity and complexity of the field of agent-based modelling and the prevalence of tools that are difficult for code-agnostic researchers to use, PerMedCoE has organized a community-driven benchmark to help map this field.
On 22 and 23 September 2022, the PerMedCoE project hosted a community benchmarking hackathon at the Barcelona Supercomputing Center (BSC). In this hybrid event, the developers of four agent-based modelling tools ran tests and simulated a variety of use cases within a time-bound contest using a predefined collection of reference datasets and assessment metrics. It was clear that even the simple tests that were run yielded different results among the tools, highlighting the need for further study of the simulation results, code, and underlying mathematics of each tool.
PhysiCell (BSC, Indiana U), Chaste (Oxford U), BioDynaMo (CERN, Sussex U) and TiSim/CellSys (INRIA, Leipzig U) participated in this hackathon with 12 people participating in person at the BSC and 10 participating online. The goal of the hackathon was to adapt the tools to the list of tests and to simulate the maximum number of use cases in this two-day exercise. All teams agreed on a set of unit tests that targeted one scale of the multiscale framework (diffusion of chemicals, mechanics of agents, agent growth and division) as well as two types of more complex use cases: a 2D monolayer and a 3D spheroid growth.
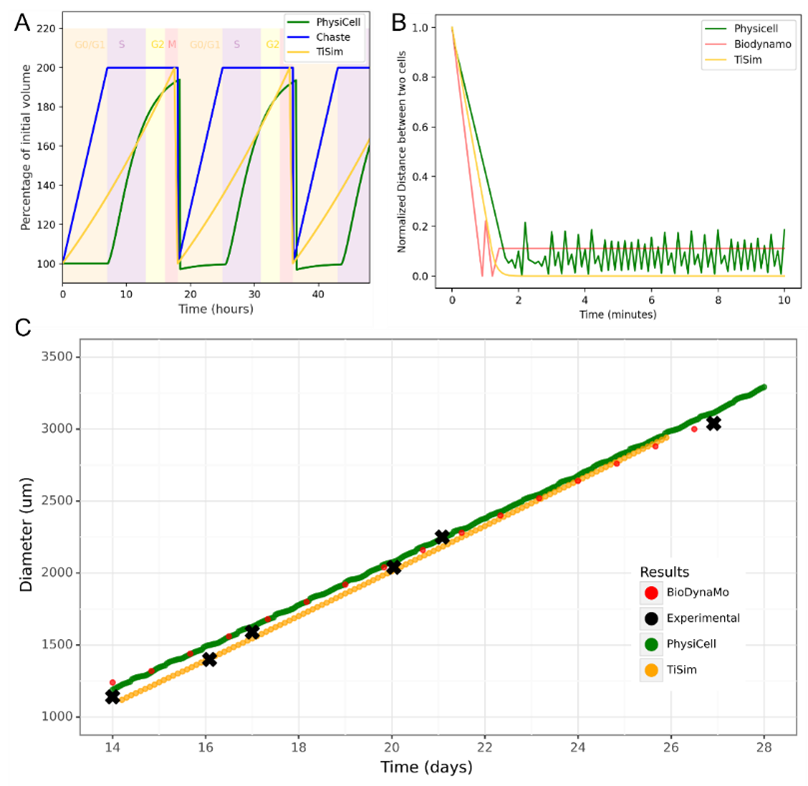
The different results among the tools were already apparent in the simple unit tests (Figure 1). Differences were seen when studying whether the tools behaved similarly when simulating a fixed-duration cell cycle model (Figure 1A) and when studying the mechanics equation of each tool by having two cells pushing each other (Figure 1B). Conversely, the tools fit well to a set of experimental growth values of a 2D monolayer growing in vitro (Drasdo and Hoehme, 2005; Hoehme and Drasdo, 2010) (Figure 1C), even though the slopes of the results of the tools were slightly different.
The results of this benchmark are available via the PerMedCoE bio.tools database, and a public website at ELIXIR’s OpenEBench platform, currently in beta, is being built to host the open methodology and the results of the benchmarks. This platform will thus serve as a base point for many other tool authors and users to add their observations and benchmarks. More information is available at the dedicated benchmark GitHub repository.
PerMedCoE also benchmarks tools from the metabolic and Boolean modelling fields and for the contextualization of signalling networks. In future benchmarks, PerMedCoE will focus on performance, runtime execution, and high-performance computing scalability tests.
Figure 1: Selected results from the agent-based modelling benchmark tests. A, Simulated cells’ volume dynamics with a cell cycle of a fixed duration. B, Simulated dynamics of the distance between the two cells pushing each other. BioDynaMo (red), Chaste





